Alison Elizabeth Murray
Molecular, ecological, microbiological and genomic researchResearch Projects
display: none
Deciphering the metagenome-encoded pathway and domain structure for Palmerolide biosynthesis.
Project Objective: To identify the Pal producing gene cluster (an antimelanoma gene product) in the microbial metagenome of the Antarctic ascidian, Synoicum adareanum. This entails assembling, annotating and mining the S. adareanum microbial metagenome. Then, a predictive bioorganic synthesis strategy will be applied to identify potential Palmerolide biosynthetic gene clusters. These predictions will be validated using single cell genome sequencing approach to screen candidate organism(s) gene content.
Project Team:
Alison Murray, DRI (lead)
Bill Baker, USF, Co-I
Patrick Chain, LANL, Co-I
Armand Dichosa, LANL, Senior Scientist
Funded by: DRI | IPA | NIH NCI Clinical and Translational R21
Advancing Nanomotion Sensor Technology to Provide Evidence for Active Life in Ocean Worlds. (NASA Cold-Tech program)
Methanogenic biodiversity and activity in Arctic and Sub-Antarctic ecosystems affected by climate change
For additional information:
Visit: http://maialenbarret.wixsite.com/methanobase
Visit: http://mars.biodiversity.aq
Building capacity in Interdisciplinary Snow Sciences for a Changing World

Dr. Murray on the Sierra Nevada crest (near Mount Lincoln) where samples were collected monthly to determine snow physical characteristics, levels of dissolved nutrients, dust, and the diversity of life in the snowpack.
For additional information: https://nasa.epscorspo.nevada.edu/nv-nasa-epscor-highlight-alison-murray-unr/
Assessing winter picoplankton distributions and carbon cycling in the North Western Antarctic Peninsula Region
The metagenomics and metatranscriptomics analyses of the Lake Vida brine microbial community reveals lifestyles at the low temperature limits of life.
The metagenomics and metatranscriptomics analyses of the Lake Vida brine microbial community reveals lifestyles at the low temperature limits of life.
Geochemistry and Microbiology of the Extreme Aquatic Environment in Lake Vida, East Antarctica. Lake Vida III: Developing a comprehensive understanding of Lake Vida – from the past to the present – life, biogeochemical function, geochemical composition and paleohistory of its microbial inhabitants.
Europa Lander Science Definition Team
Prof. Murray was co‐chair of a team of 21 scientists that set out to establish the science objectives and investigations of NASA’s goals for the first landed mission to the surface of Europa, which are: 1) To search for evidence of life on Europa; 2) To assess the habitability of Europa via in situ techniques uniquely available to a lander mission; and 3) Characterize surface and subsurface properties at the scale of the lander to support future exploration. Europa, a moon of Jupiter is the closest ocean world to Earth, and holds promise as being potentially habitable. The constraints of such a mission are severe – the Europan surface is bombarded by radiation from Jupiter, has no atmosphere, and is a frigid ‐160 oC.
View report:
https://solarsystem.nasa.gov/docs/Europa_Lander_SDT_Report_2016.pdf
Wikipedia entry:
https://en.wikipedia.org/wiki/Europa_Lander_(NASA)
Past Research Projects
Antarctic Microbial Biology
display: none
Astrobiology of Icy Worlds: Habitability, Survivability, and Detectability
Project Objective: The primary goal in the Astrobiology of Icy Worlds Investigation is to advance our understanding of the role of ice in the broad context of astrobiology through combined laboratory, numerical, analytical, and field investigations. The Icy Worlds team is pursuing this goal through four major investigations; the habitability, survivability, and detectability of life of icy worlds coupled with “Path to Flight” technology demonstrations.
As the team searches for signatures for life by “following the ice,” such questions have emerged as can life emerge and thrive in a cold, lightless world beneath hundreds of kilometers of ice? And if so, do the icy shells hold clues to life in the subsurface environment? These questions are the primary motivation of these science investigations.
More information:
The NASA Jet Propulsion Laboratory: https://www.jpl.nasa.gov/
The NASA Astrobiology Institute:
https://nai.nasa.gov/directory/murray-alison/
Funded by: DRI | NASA Jet Propulsion Laboratory
IPY: Bacterioplankton Genomic Adaptations to Antarctic Winter
Project dates: 06/15/2007 – 05/31/2010
Funded by: National Science Foundation
Project Description:
The primary objectives of this program are:
- Describe the differences in diversity and genomic content between austral winter and summer bacterioplankton communities. This will allow us to test hypotheses concerning (i) the diversity of bacterioplankton between the Antarctic coastal ecosystem and other lower latitude systems (ii) the functional capacity revealed through a genome content survey and (iii) that there are adaptations (e.g. in amino acid modification) inherent in the Polar bacterioplankton environmental genome that are season-independent.
- Investigate the winter-time bacterioplankton growth and cellular signals (mRNA and proteins expressed) in order to understand the specific adaptations and keys to survival. Hypotheses here will test that (i) Bacteria respond to the onset of Austral winter by transitioning into the starvation-survival mode and (ii) Planktonic marine crenarchaeota possess adaptations for enhanced growth and survival in the Austral winter, enabling them to increase in abundance and become a dominant part of the picoplankton community.
Through support from the CSP we have sequenced 8 bacterial SSU rRNA gene libraries spanning the annual cycle totaling over 5500 sequences, 2 eukaryal SSU rRNA gene libraries from austral summer and winter (~1400 sequences), and one archaeal library from austral winter (~700 sequences).
Two large contig (~40kb) fosmid metagenomic libraries have also been created with 10K clones each, and the ends of these fosmid clones have been sequenced resulting in ~ 37,000 end sequences. We also have selected a number of these clones for complete sequencing.
Free-drifting icebergs as proliferating dispersion sites of iron enrichment, organic carbon production and export in the southern ocean
Project period: 3/2009 – 2011
Funded by: NSF Office of Polar Programs
Project Description: The Murray group (B-014) component of the iceberg project is to address the following question: What is the relationship between Fe and nutrient distributions associated with free-drifting icebergs and the organic carbon dynamics of the ice-attached and surrounding microbial communities?
We will study the organic carbon cycling abilities of the picoplankton. We hypothesize that microbial plankton will respond to changes in micronutrient regime and water column structure imposed by the iceberg. These changes will result in a greater amount of carbon production and export to the deep sea, in addition to supplying a system capable of rapid DOC utilization. The primary objective is to integrate measured microbial biomass, activity and diversity in the iceberg zone of influence (IZI) in comparison to unaffected, control areas. The second objective will be to determine the linkages between primary and secondary production in the IZI and control areas, and how the activities are related to community structure, trophic dynamics and sedimentation rates. Thirdly, microbial community fingerprints will be used as bio-signatures of water column mixing to determine biotic influences of iceberg-associated upwelling.
Subsurface ice and brine sampling: life detection and characterization in the McMurdo Dry Valleys using an ultrasonic gopher.
PI/COPI: Peter Doran, UIC (PI); Chris Fritsen, Alison Murray, Yoseph (Yosi) Bar-Cohen, JPL; Fabein Kenig, UIC; Chris McKay, NASA AMES.
Funded by: National Science Foundation Office of Polar Programs
Collaborators: John Priscu, Montana State University
Keywords: astrobiology, subsurface ice, brine, McMurdo Dry Valleys, Antarctica, ultrasound, Lake Vida, drilling
Project Description: Evidence from ice cores and ground penetrating radar suggested that the previously thought to be completely frozen Lake Vida has a saturated brine lake underlying the 60 ft. ice cover. The goal of this project was to sample the subsurface brine in Lake Vida using a new drilling technology. This brine is one of the most extreme aquatic habitats on the planet – and whether life exists there is still unknown. Along with Chris Fritsen at DRI and other colleagues from University of Illinois we embarked on a field expedition in December 2005 to sample the brine. We did not enter the lake, but sampled brine that invaded the deep hole we drilled. At McMurdo Station we ran assays to characterize the microbial activity in this rather inhospitable brine (-13C, 6X salinity of seawater, and dissolved nutrients and metals that may prohibit growth). Now at DRI we are performing DNA sequencing of bacterial genes to determine the identities of the life inhabiting the brine collected from the ice cover.
Project Website: NASA Astrobiology Science and Technology for Exploring Planets (ASTEP) Program
Genome Sequencing of two Antarctic bacteria – a marine Actinobacterium, str. PHSC20C1 and a sea-ice associated bacterium, Polaribacter irgensii, 23-P.
Project Period: 9/2004
Funded by: The Marine Microbiology Initiative Program at the Gordon and Betty Moore Foundation
Project Description:
The understanding of what features of microorganisms have led to their success in cold, subzero environments is currently unknown – however the genome of organisms that thrive in these environments promises to unravel information concerning their physiological adaptations, evolutionary history, and overall genome content useful in comparative genomic studies with organisms from lower latitudes. The genomes are presently in high draft format, have been automatically annotated using NCBI’s PGAAP and were submitted to GenBank in March 2006 (PHSC20C1: AAOB00000000, Polaribacter irgensii 23-P: AAOG00000000).
Gene Expression in Extreme Environments: Extending Microarray Technology to Understand Life at its Limits
Project period: 3/2001 – 2/2004
Funded by: National Science Foundation, Life in Extreme Environments Program
Project description: This project has aimed to develop the technology for studying global gene expression in microorganisms sampled directly from the environment, our study area was Antarctic Peninsula coastal waters, where temperatures are constantly cold (-2 to -1o C) . We are working on characterizing the diversity of the coastal Antarctic marine bacterioplankton, and have sequenced a number of environmental genomic DNA fragments cloned directly from the environment.
Environmental genomics of Antarctic marine bacterioplankton using a large-contig sequencing approach
Project period: 1/2006
Funded by: DOE – Joint Genome Institute Community Sequencing Program
Project description: In order to get a much more enhanced and detailed picture of marine bacterioplankton genomes the Joint Genome Institute will sequence environmental genomic DNA from late winter and summer genomic libraries sampled from the waters off-shore of Palmer Station, Antarctic Peninsula. We still know little of nature of the differences in communities inhabiting surface waters during summer and winter – though they are quite different in composition (~ 60% different). Both libraries will be end-sequenced producing 34,000 single read sequences. Then using comparative genomic approaches we may be able to distinguish the genomic features that make the bacterioplankton communities distinct between these two different times of year. We then will select 150 large insert clones to sequence from each library which will provide more detailed information concerning this subset of the whole representing both unique, and commonly represented members of the community.
More information: CSP 2006 Sequencing plans
Hydrothermal Microbial Biology
display: none
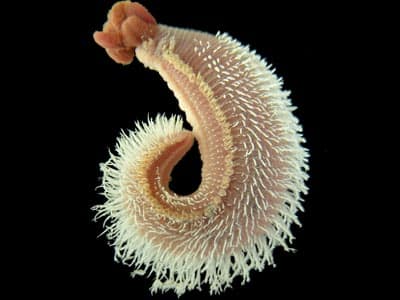
Vent Epibiont Environmental Genome (VEEG) -A meta-genome level analysis of an extreme microbial symbiosis
Project period: 1/2002
Funded by: NSF – Biocomplexity GENEN program
Project description: In collaboration with Dr. Craig Cary, University of Delaware and others, this interdisciplinary project’s goal has been to reconstruct the metabolism of a consortia of bacteria that live symbiotically with a polychaete worm which inhabits the walls of hydrothermal vent chimneys at depths from 2-3000 meters under the sea’s surface. The approach for metabolic reconstruction has involved sequencing a large amount of DNA from this consortia and implementing a multi-step analysis process to sort through the sequence to identify genes and pathways that are encoded in it. We have participated in 2 ocean cruises and used the submersible Alvin to sample the worms (and their RNA) and study the environmental characteristics of their toxic, heat-laden habitat. Now that hypotheses are being formed regarding the metabolism of the symbionts we will construct DNA microarrays in the near future to see if the pathways suspected to be important are actually expressed. We have learned that the genomic complexity of the symbiont a community is much higher than originally thought – though we have been successful in reconstructing metabolic pathways for many of the major biogeochemical cycles that are occurring in this environment.
Geobiology and the emergence of terraced architecture during carbonate mineralization
Project period: 9/2002
Funded by: NSF – Biocomplexity: Coupled Biogeochemical Cycles
Project description: Carbonate terraces are a pervasive geological form on this planet, and perhaps other planets. This project, led by Bruce Fouke of the University of Illinois, has taken place at Mammoth Hot Springs, Yellowstone National Park. The aim has been to decipher the origins and evolution of carbonate terraces as a function of the biological, chemical and physical regime in hot spring systems. The Murray group has participated in four key aspects of this field-oriented geobiolgy research program. (i) They have described the community structure of bacteria over geological facies transitions and in association with specific crystalline features in the hot spring system. The community structure varies closely with the geological facies model based on chemical/physical boundaries in the system. (ii) An examination into the phylogenetic diversity of archaeal thermophiles in the hot spring vent pool has shown that there are several forms of archaeal life that appear to be abundant there, one that appears to be quite distinct from anything known, and the another that forms a very deep-branching lineage (Korarchaeota) that has no cultivated members. (iii) They have surveyed carbon fixation pathways (and genes involved in them) that are active in photosynthesis in the hot spring system where temperatures exceed 45C. (iv) In fall 2005, they participated in an experimental field study aimed to study carbonate mineralization in hot spring waters with microbes present, with killed microbes, and with the microbes removed by filtration. They are working to the identity of organisms associated with primary mineralization and biofilm development events, while the Illinois groups are working on comparing the quantity and form of minerals formed in each of the three treatments.
More information:
DRI News: Seeking the origin of Yellowstone’s Travertine Terrace Formation: Are the bugs involved? DRI Summer Newsletter 2003
CONTACT
Alison Murray, Ph.D.
Alison.Murray@dri.edu
LAB LOCATION
Desert Research Institute
2215 Raggio Parkway
Reno, NV 89512
DIVISION
Earth & Ecosystem Sciences